Integration
Function Interface
The shared memory versions of algorithms are implemented inside relevant functions using OpenMP or C++::threads depending on the function’s specification. The following sub-sections will provide an overview of integrating new or existing shared memory database peptide search algorithms with HiCOPS parallel library.
Database Indexing
The peptide database segments (length by length) are indexed in a loop inside the function DSLIM_Construct(Index *) using OpenMP. The index data structure passed to this function is as follows:
struct Index
{
uint_t pepCount; // global count of normal peptides in this segment
uint_t modCount; // global count of modified peptides in this segment
uint_t totalCount; // global pepCount + modCount
uint_t lclpepCnt; // number of normal peptides in this segment in this MPI task
uint_t lclmodCnt; // number of modified peptides in this segment in this MPI task
uint_t lcltotCnt; // lclpepCnt + lclmodCnt in this MPI task
uint_t nChunks; // number of internal chunks in this segment in this MPI task.
uint_t chunksize; // size of internal chunk in this segment in this MPI task
uint_t lastchunksize; // size of the last chunk in this segment in this MPI task
PepSeqs pepIndex; // contains a list of peptide sequences
pepEntry *pepEntries; // lcltotCnt sized list of database entries (first lclpepCnt: normal, last lclmodCnt: variants).
SLMchunk *ionIndex; // nChunk sized list of spectra index items. Refer to: https://ieeexplore.ieee.org/abstract/document/8983152
}
MS/MS Data Preprocessing
A partition of experimental MS/MS data are pre-processed inside DSLIM_InitializeMS2Data() using OpenMP at each MPI task. This function constructs file and tag index objects: MSQuery for the complete MS/MS dataset at each MPI task. The MSQuery class has a built-in MS2 file parser as well. The MSQuery class can thus be updated as needed for data pre-processing. We recommend pre-processing only a chunk of dataset at each MPI task.
static status_t DSLIM_InitializeMS2Data()
{
// dataset files
int_t nfiles = queryfiles.size();
// create a list of MSQuery objects
ptrs = new MSQuery*[nfiles];
#ifdef USE_OMP
#pragma omp parallel for schedule (static)
#endif/* _OPENMP */
for (auto fid = 0; fid < nfiles; fid++)
{
// create a new object for each file
ptrs[fid] = new MSQuery;
// Pre-Process a dataset file with the object
ptrs[fid]->InitQueryFile(&queryfiles[fid], fid);
}
/* ALREADY IMPLEMENTED CODE HERE */
return status;
}
Database Search & Scoring
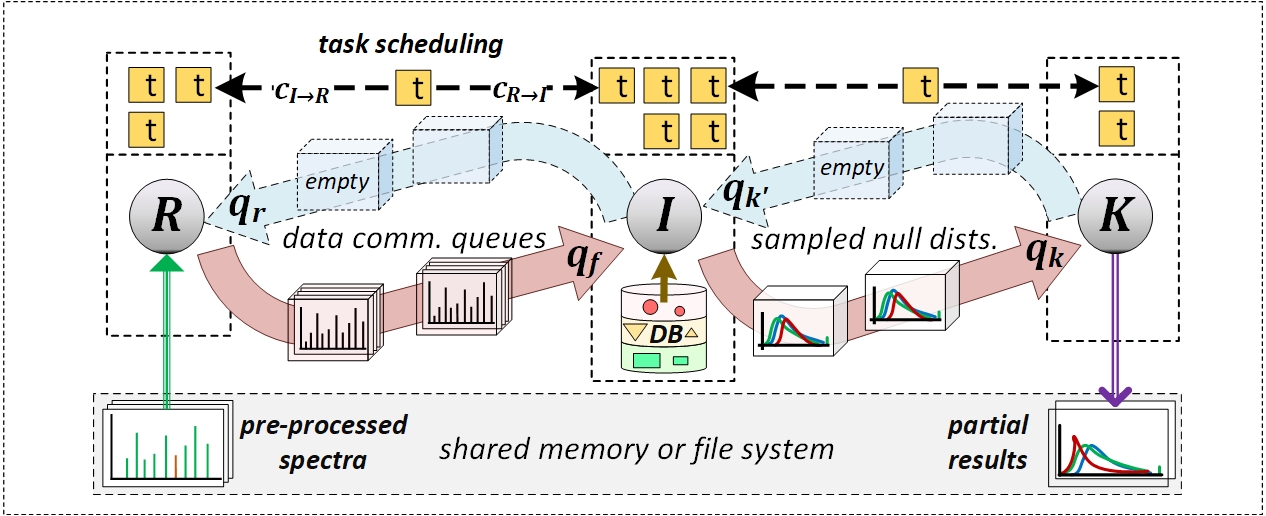
The database search (scoring) superstep in HiCOPS is implemented through 3 parallel sub-tasks, R, I, and K. The parallel sub-tasks work in a producer consumer model with variable number of parallel threads dynamically managed between them. The sub-tasks R and K are implemented using std::thread while the sub-task I has been implemented via OpenMP.
- The sub-task R reads batches of pre-processed experimental data (using the batch index) and queues them into q_f.
- The sub-task I extracts each batch from q_f, executes the partial database peptide search producing intermediate results that are queued into q_k. The consumed data buffers are recycled to sub-task R through q_r.
- The sub-task K reads batches of intermediate results from q_k and writes them to the shared memory or file-system. The consumed memory buffers are returned to the sub-task R through q’_k.
A schematic setup of parallel sub-task setup along with task-scheduling and queues is shown in the following figure. For more details, please refer to the original research paper here.
Relevant Functions
- The sub-task R executes the following function using
std::threadparallel model.
void DSLIM_IO_Threads_Entry();
- The sub-task I executes the function using
OpenMPparallel fashion.
status DSLIM_QuerySpectrum(Queries *batch,
Index *index,
uint_t total_index_segments);
- The sub-task K executes the following function using
std::threadparallel model.
void DSLIM_FOut_Thread_Entry();
Statistical Scoring
A partition of the intermediate results produced by the HiCOPS are assembled at each MPI task inside the function: DSLIM_Score::CombineResults() using OpenMP. The expeRT class can model the survival functions using either tail-fit or logWeibull fit. The expeRT class also implements Savitzky-Golay data smoothing, curve fitting algorithms and light-weight py-numpy like vectors which can be used as is or modified to build most statistical scoring techniques.
False Discovery Rate (FDR)
The false discovery rate (FDR) calculation has not yet been implemented in HiCOPS source and you are welcome to contribute in the development of this feature. The FDR can be implemented as a separate parallel binary application.
Active Development (Future)
We are actively developing a new C++ template meta-programming based interface for HiCOPS allowing simple modular application following the build your own workflow philosophy. The new interface will be similar to LLVM and Timemory allowing users to easily build and customize their workflows without having to parallelize (even with OpenMP or threads) or directly interact with the core library.