Features
This various features provided by the HiCOPS include:
Parallel Core Design
The HiCOPS parallel core design is based on the Bulk Synchronous Parallel (BSP) model building the database peptide search workflow using four supersteps. The algorithmic work in each superstep is executed in asynchronous parallel fashion and synchronization is performed between supersteps, if required. The first superstep partitions the massive database in a load balanced fashion among parallel MPI tasks. The second superstep preprocesses batches of experimental MS data and creates an index. The third superstep performs the partial database peptide search storing the intermediate results on the shared file system. The final superstep assembles the intermediate results yielding complete results. A high-level graphical design of HiCOPS superstep model is shown in the following figure.
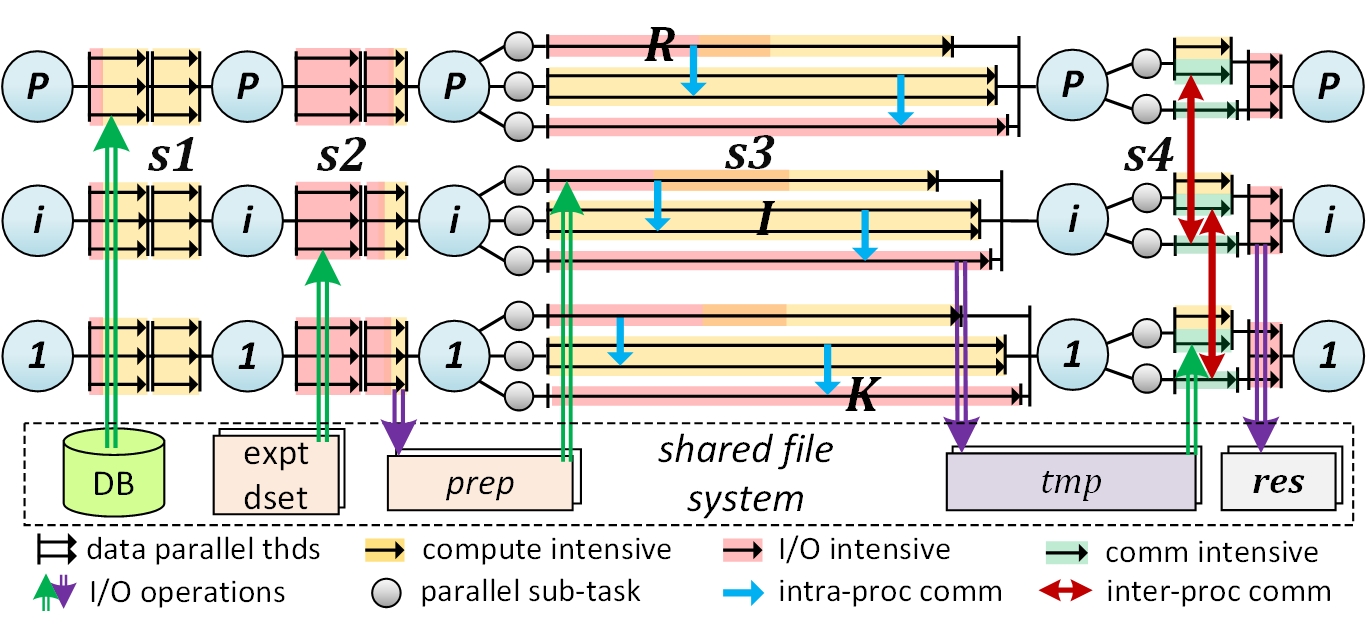
Several optimization techniques including buffering, sampling, overlapped computations and communications and task scheduling are implemented to aid in optimal parallel performance. Further, the core library packs parallel versions of many core algorithms and the essential boiler-plate code for quicker development of new tools based on the discussed parallel design. Read more about the HiCOPS parallel design, its mathematical analysis and optimizations in our original research paper here.
Performance
The proposed parallel design along with appropriate optimizations allow maximum utilization of underlying resources with minimal overheads enabling ultra-fast peptide search speeds. The highlights of performance evaluation results include:
- Speedup vs Other tools: More than 100x against all tools and up to 420x for large workloads compared to MSFragger.
- Parallel Efficiency: 75-85% when the workload size is sufficiently large for up to 72 parallel nodes (x24 = 1728 cores).
- Load Imabalance: Less than 10%.
- Roofline: Super-linear speedups with increasing nodes when the problem size is large.
- Comm/Compute Ratio: Less than 5%.
Read more about HiCOPS results, scalability and performance in our original research paper here.
Performance Analysis Support
Performance analysis and instrumentation support has been provided at multiple levels via Timemory toolkit using C++14 extern templates allowing custom instantiation of only the required instrumentation components resulting in reduced compile times. Please refer to Timemory README on GitHub more information on the instrumentation interface. The instrumentation can be modified at pre-runtime using environment variable explained here.
Command-line Tools
HiCOPS provides several command-line tools for auxiliary purposes including database and experimental data preparation and extraction, post-processing results and runtime support. Please refer to Tools for detailed information on tools.
Integratability
The software design allows any existing or novel database peptide search algorithms such as data indexing algorithms, search algorithms, scoring algorithms, post-processing, data analytics algorithms to be easily integrated with HiCOPS. For more information on integration with HiCOPS, please refer to the Integration document.